![]()
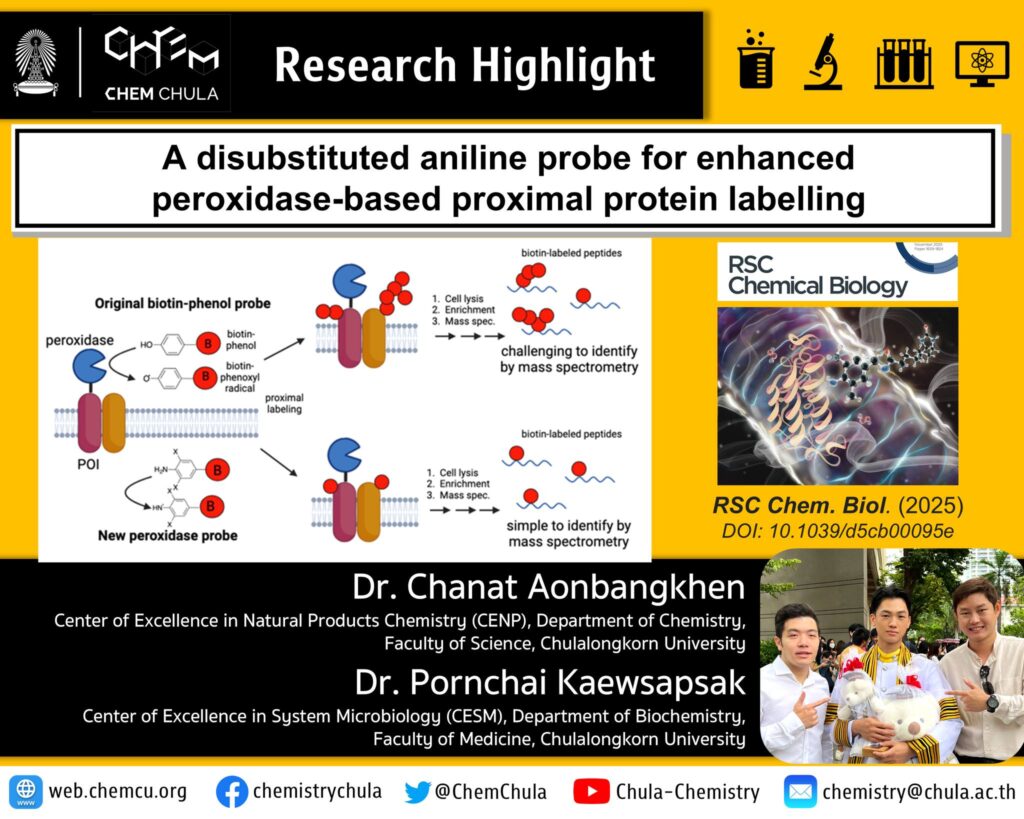
![]() Proteins are biomolecules essential for cellular functions, including cell signaling and regulation. Protein misfolding or mislocalisation can result in various diseases. Peroxidase-mediated proximity labelling has emerged as a powerful tool for studying subcellular proteome and protein–protein interactions. However, the traditional probe, biotin-phenol, suffers from limitations including low protein enrichment efficiency, and the formation of oxidised and polymerised products, complicating the downstream analysis.
Proteins are biomolecules essential for cellular functions, including cell signaling and regulation. Protein misfolding or mislocalisation can result in various diseases. Peroxidase-mediated proximity labelling has emerged as a powerful tool for studying subcellular proteome and protein–protein interactions. However, the traditional probe, biotin-phenol, suffers from limitations including low protein enrichment efficiency, and the formation of oxidised and polymerised products, complicating the downstream analysis.![]()
![]() To address these challenges, a novel probe, N-(4-amino-3,5-dimethylbenzyl)desthiobiotinamide (DBA-Me), for protein labelling in living cells was developed, by Dr.Chanat Aonbangkhen, and Mr.Nattavorapon Tantisasiratin, the Master’s student researcher in the 4+1 program of the Department of Chemistry, as well as the faculty members of the Department, in close collaboration with Dr.Pornchai Kaewsapsak and his team from the Department of Biochemistry, Faculty of Medicine, Chulalongkorn University, as well as other faculty members in multiple other institutes in Thailand, such as the National Center for Genetic Engineering and Biotechnology (BIOTEC), NSTDA, and Chulabhorn Graduate Institute (CGI)- Chulabhorn Royal Academy(CRA).
To address these challenges, a novel probe, N-(4-amino-3,5-dimethylbenzyl)desthiobiotinamide (DBA-Me), for protein labelling in living cells was developed, by Dr.Chanat Aonbangkhen, and Mr.Nattavorapon Tantisasiratin, the Master’s student researcher in the 4+1 program of the Department of Chemistry, as well as the faculty members of the Department, in close collaboration with Dr.Pornchai Kaewsapsak and his team from the Department of Biochemistry, Faculty of Medicine, Chulalongkorn University, as well as other faculty members in multiple other institutes in Thailand, such as the National Center for Genetic Engineering and Biotechnology (BIOTEC), NSTDA, and Chulabhorn Graduate Institute (CGI)- Chulabhorn Royal Academy(CRA).![]()
![]() Western blot analysis demonstrated efficient labelling of bovine serum albumin in vitro. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) data confirmed the formation of one-to-one adducts from the in vitro labelling reaction. Notably, DBA-Me also permits APEX2-mediated labelling within the mitochondrial matrix of HEK293FT cells and demonstrated improved recovery of labelled proteins after streptavidin enrichment compared to the conventional biotin-phenol (BP) probe, highlighting its superior potential application in cellulo. This facilitates peroxidase-mediated proximity labelling applications in subcellular localisation of proteins, and protein structures, with broader implications for understanding cellular processes and disease mechanisms. Moreover, this novel probe (DBA-Me) also exhibited labelling activity towards nucleic acids, which might be used to study other biomolecules in the near future.
Western blot analysis demonstrated efficient labelling of bovine serum albumin in vitro. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) data confirmed the formation of one-to-one adducts from the in vitro labelling reaction. Notably, DBA-Me also permits APEX2-mediated labelling within the mitochondrial matrix of HEK293FT cells and demonstrated improved recovery of labelled proteins after streptavidin enrichment compared to the conventional biotin-phenol (BP) probe, highlighting its superior potential application in cellulo. This facilitates peroxidase-mediated proximity labelling applications in subcellular localisation of proteins, and protein structures, with broader implications for understanding cellular processes and disease mechanisms. Moreover, this novel probe (DBA-Me) also exhibited labelling activity towards nucleic acids, which might be used to study other biomolecules in the near future. ![]()
![]()
- Pornchai Kaewsapsak
- Nattavorapon Tantisasirat
- Sucheewin Krobthong
- Peeraphan Compiro
- Ariya Khamwut
- Kidakarn Ratchakitprakarn
- Naphat Chantaravisoot
- Kriangsak Faikhruea
- Withsakorn Sangsuwan
- Medena Noikham
- Worawan Bhanthumnavin
- Tirayut Vilaivan
- Sunchai Payungporn
- Yodying Yingchutrakul
- Watthanachai Jumpathong
- Chanat Aonbangkhen